George here, with a brief summary of our revised preprint; the first native RNA sequence of SARS-CoV-2 using @nanopore, and a data-driven inference of genetic features and evolutionary rate. 1/12
https://www.biorxiv.org/content/10.1101/2020.03.05.976167v2.full.pdf">https://www.biorxiv.org/content/1...
https://www.biorxiv.org/content/10.1101/2020.03.05.976167v2.full.pdf">https://www.biorxiv.org/content/1...
Ko tenei tuhinga e wehewehe ana i nga waahanga o te mate COVID-19, a ka awhina pea ia tatou ki te whakatau i tenei wero 2/12
The SARS-CoV-2 materials for native RNA sequences were derived from the first SARS-CoV-2 culture achieved outside of China, now including sequences from both cell material and purified virions. 3/12 https://twitter.com/TheDohertyInst/status/1222232959375347713?s=20">https://twitter.com/TheDohert...
The RNA generated from SARS-CoV-2-infected cellular material was expected to include all viral transcripts actively expressed by the virus, with a range of methods being used to define major transcripts. 4/12
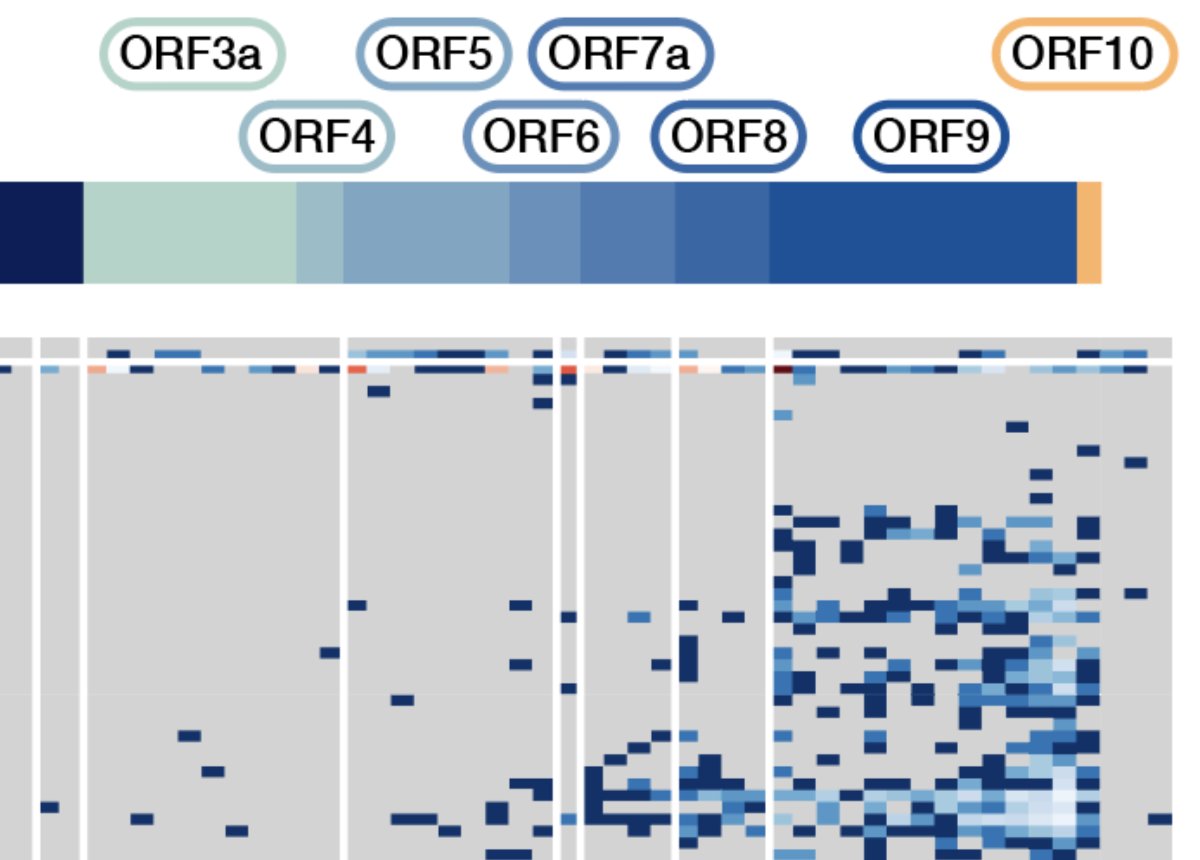
The defined SARS-CoV-2 transcripts are close to completely absent in the purified virion data, with the RNA generated here including genomic RNA as the major component. 5/12
Interestingly, one of the annotated open reading frames in SARS-CoV-2 does not appear as a major transcript in this dataset, an observation also made by Davidson et al. and Kim et al. in further direct RNA projects. 6/12 https://www.biorxiv.org/content/10.1101/2020.03.12.988865v2">https://www.biorxiv.org/content/1...
The expected peptide encoded by ORF10 in SARS-CoV-2 does not appear in early proteomic data , further supporting the hypothesis that the sequence annotated as ORF10 may not have a protein coding function in SARS-CoV-2. 7/12
Other features identified by the native RNA sequence include probable RNA modifications, a handful of these being identified at consistent sequences in SARS-CoV-2 transcripts 8/12
Aspects of the early evolution of SARS-CoV-2 are also described, including estimations of the evolutionary rate and time-to-most-common-recent-ancestor for the pandemic, thought to be in late November 2019 based on our phylodynamic methods 9/12
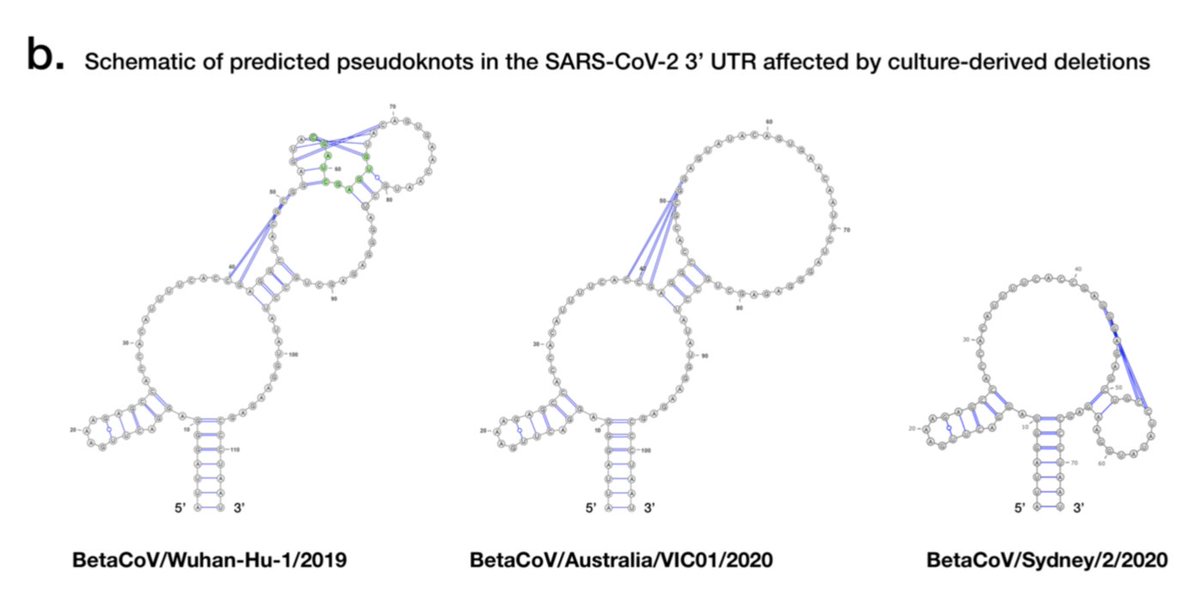
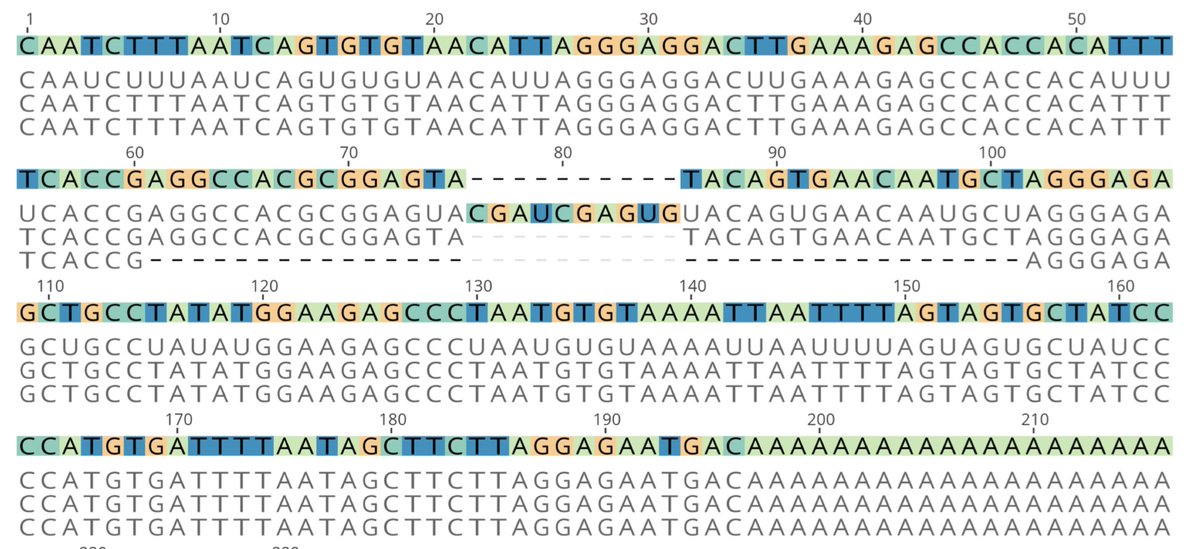
A small number of culture-derived SARS-CoV-2 genomes are also shown to carry deletions at the 3’ UTR; the UTR including areas thought to have roles in regulating viral RNA synthesis 10/12
As mentioned, other groups have made further descriptions of SARS-CoV-2 RNA through direct sequencing, including fantastic work by Davidson et al. @CMMBristol and Kim et al. @hyeshik. Each of us have also made our data publicly available 11/12
Many thanks to the collaborators on the project, with at least half of the co-authors being involved in broader aspects of the local response to COVID-19 here at Melbourne. Hugely grateful @sebduchene @lachlanjmc @dnrawl @Miranda_E_Pitt @Nick_E_Scott and others 12/12

 Read on Twitter
Read on Twitter