1/6 Our #SARSCoV2 PCR comparison is now online as a preprint! We show that PCR assays are able to detect SARS-CoV-2, but with different detection limits and variable ability to differentiate between true negatives and positives at low RNA concentrations. https://www.medrxiv.org/content/10.1101/2020.03.30.20048108v1.article-metrics">https://www.medrxiv.org/content/1...
2/6 We standardized PCR reagents and conditions to make a fair comparison between nine primer-probe sets from 4 commonly used assays. We determined analytical sensitivity by spiking SARS-CoV-2 RNA into mock extracts from negative NP swabs (collected in 2017).
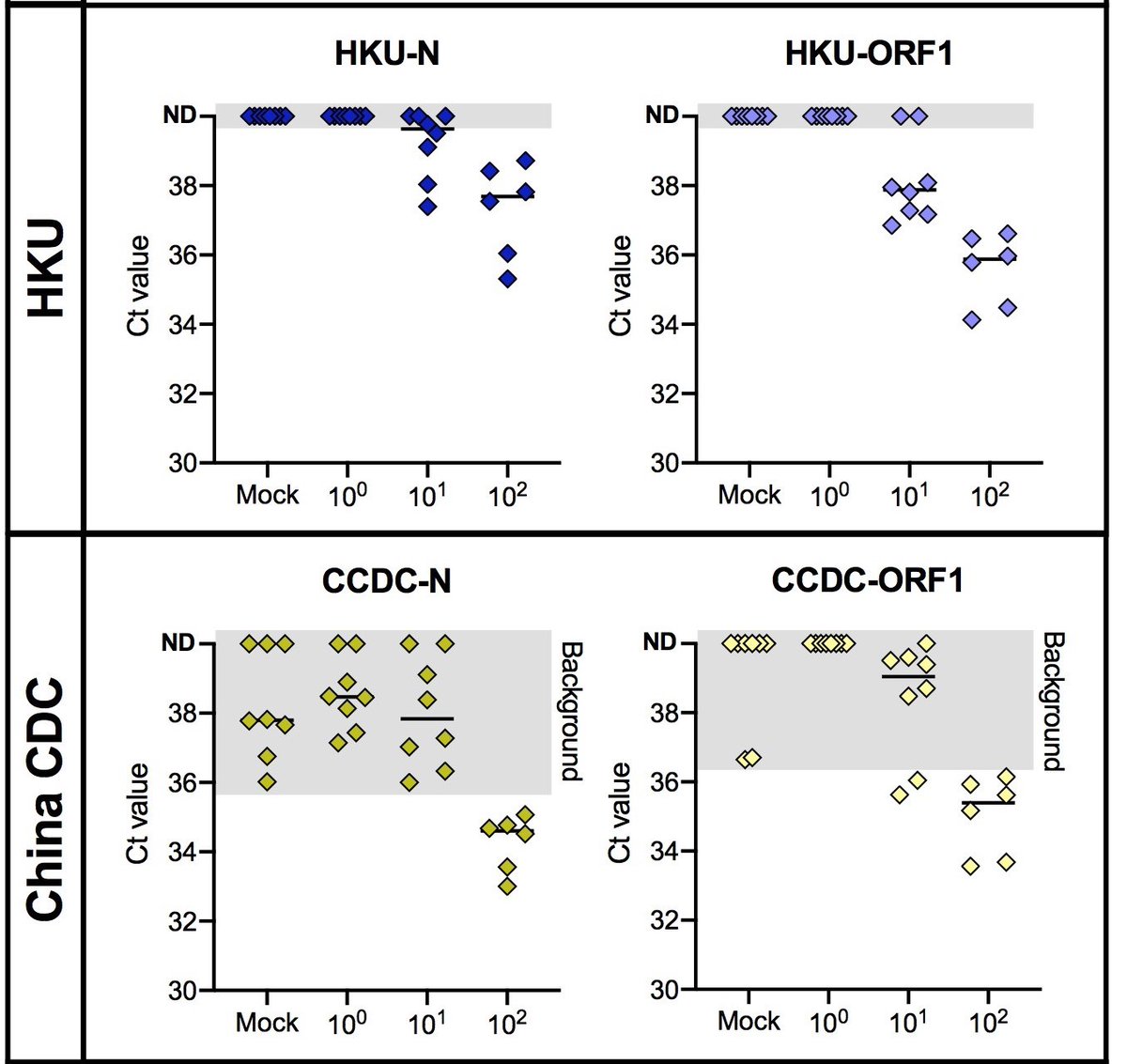
3/6 We found that the CDC assay works well, but with background amplification by N2 and N3 (now excluded from assay), which may lead to inconclusive results.
4/6 The primer-probe sets developed by Hong Kong University (HKU) had no background amplification and were very sensitive. In contrast, the China CDC primer-probe sets had high background amplification which may lead to false positive results.
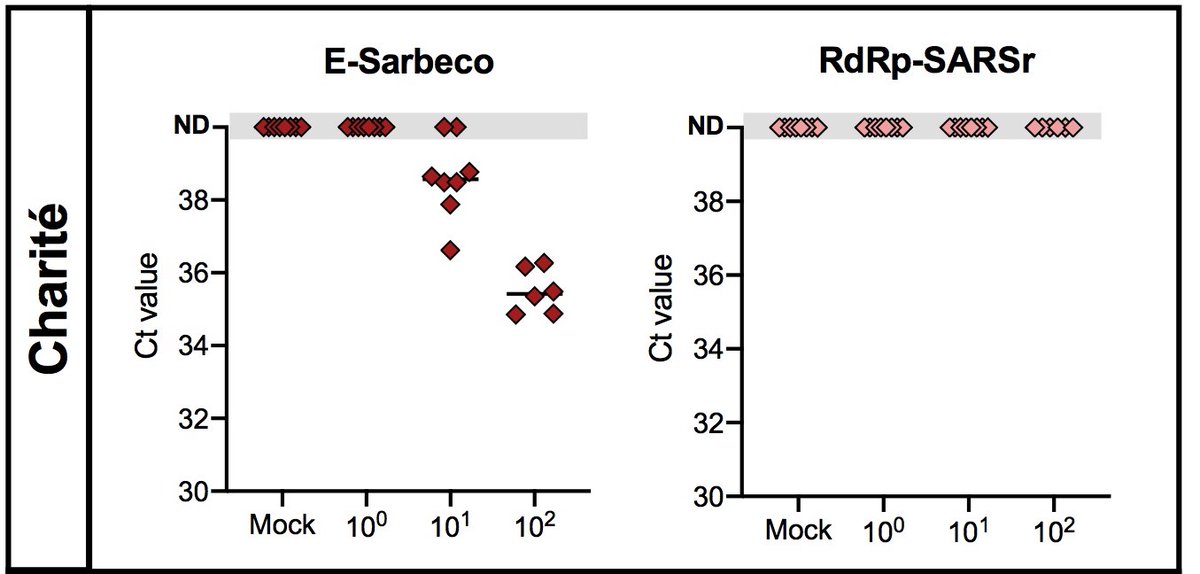
5/6 The E-Sarbeco primer-probe set developed by the Charité Universitätsmedizin Berlin had a high analytical sensitivity, but their confirmatory assay (RdRp-SARSr) had low sensitivity. This is likely due to a mismatch in the reverse primer.
6/6 Our findings will help other labs to understand the limitations of commonly used primer-probe sets. Importantly, labs need to locally validate sensitivity and positive-negative cut-off values as these may be different for other PCR kits and thermocyclers.

 Read on Twitter
Read on Twitter