There is a great data portal from @NCBI, but only had ~173 genomes and ~41 raw sequencing read data sets. https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/">https://www.ncbi.nlm.nih.gov/genbank/s...
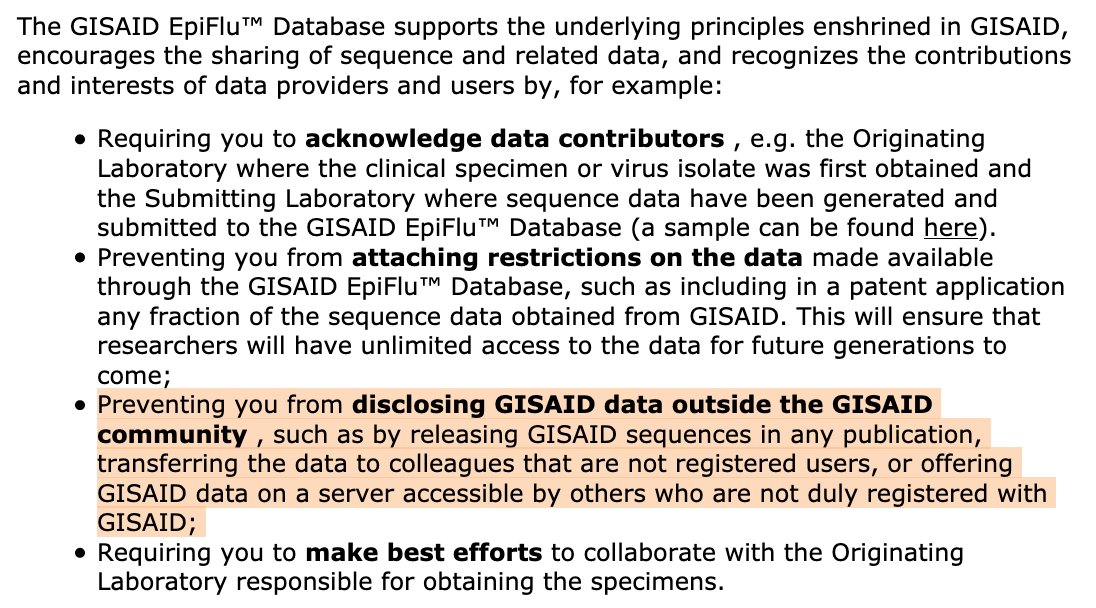
Meanwhile @nextstrain is getting their data from http://gisaid.org"> http://gisaid.org which has much more data, ~700 strains, but onerous restrictions on data use and sharing, in particular does not allow sharing any sequence data.
This is a real impediment to rapid, collaborative data analysis including our efforts to make transparent, reusable and reproducible analysis pipelines for outbreak analysis ( https://covid19.galaxyproject.org/ ">https://covid19.galaxyproject.org/">... #usegalaxy).

 Read on Twitter
Read on Twitter