I& #39;m fed up of having to make these molecular biology arguments about common ancestry over and over.
A thread
A thread
Molecular evidence includes:
1. Chromosomal similarities
2. ERV insertions at exactly homologous locations in primate genomes
3.Gross sequence homology
4. Nested hierarchies of shared variants
1. Chromosomal similarities
2. ERV insertions at exactly homologous locations in primate genomes
3.Gross sequence homology
4. Nested hierarchies of shared variants
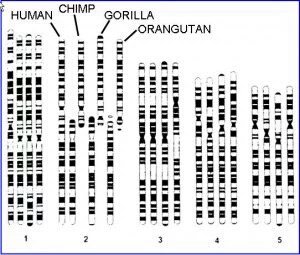
1. Human chromosome 2 is a fusion of 2 ancestral chromosomes
Not only do we see the immediate alignment of chromosome bands but DNA sequencing across the junction confirms the fusion
Plus synteny (same genes in the same order) across other chromosomes
Not only do we see the immediate alignment of chromosome bands but DNA sequencing across the junction confirms the fusion
Plus synteny (same genes in the same order) across other chromosomes
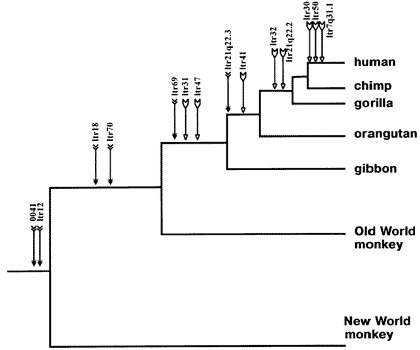
2. ERV insertion points.
These are inherited in certain lineages in a nested manner.
The chances of these insertions doing this by chance alone are infinitesimal
(See point 4)
These are inherited in certain lineages in a nested manner.
The chances of these insertions doing this by chance alone are infinitesimal
(See point 4)
Gross sequence homology is powerful evidence that sequences are shared as a result of common ancestry.
For those who say this is design, why are there any differences at all? Doesn& #39;t the designer know his best sequence? Why are there more differences in the 3rd base of codons?
For those who say this is design, why are there any differences at all? Doesn& #39;t the designer know his best sequence? Why are there more differences in the 3rd base of codons?
In addition, common design would imply that common function leads to similarity of sequence. So the prediction would be that genes for penguin flippers would more closely resemble those for seal flippers than those for hummingbird wings. This is not what we observe
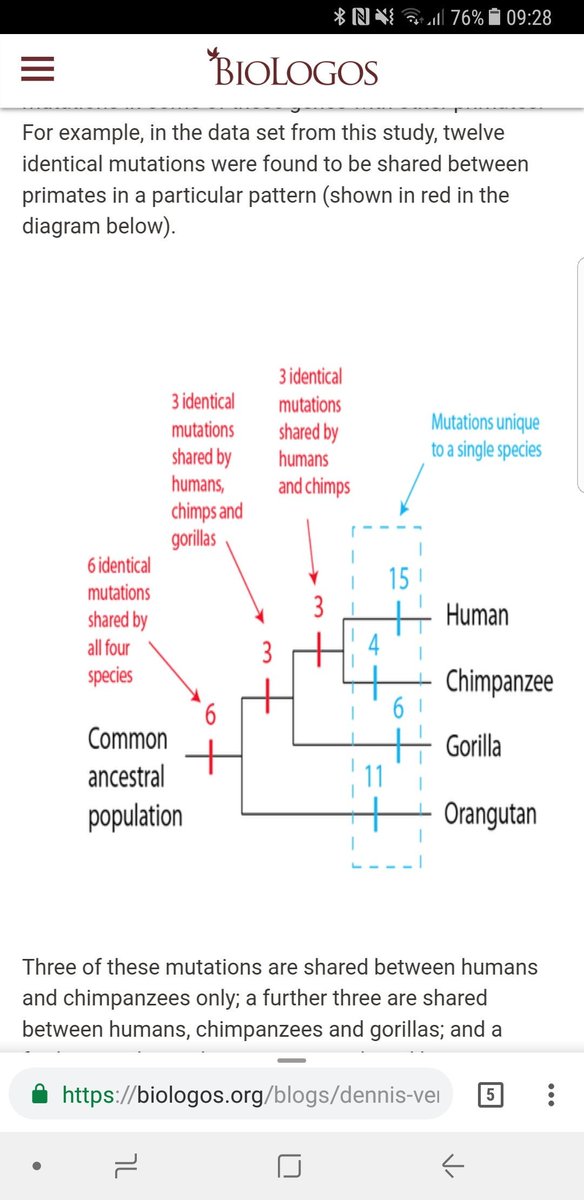
4. Nested hierarchy of variants. In the study below, there is a clear pattern of nesting. Bear in mind these are pseudogenes that produce no viable protein. There is no functional reason for these variants to be shared.
Another aspect is that the 3rd base of degenerate codons (changes which will not affect protein sequence, hence function), ALSO show this same pattern of nesting. Strong evidence of common descent rather than functional constraints/convergence
Similarly, the GULO pseudogene is mutated in EXACTLY the same way in one particular lineage of primates but not in others. Common ancestry explains this while no other hypothesis comes close
/Ends

 Read on Twitter
Read on Twitter