Paper paper! We investigated what genes malaria parasites need to develop in the liver using barcode-sequencing. Exciting collaboration led by @rrlimenitakis @bushell_lab and @AnushChP. https://authors.elsevier.com/sd/article/S0092867419311808">https://authors.elsevier.com/sd/articl...
We had previously applied barseq-based reverse-genetics to the stages Plasmodium berghei that grow in the blood, and found that an unexpectedly large proportion of genes are essential in this phase of parasite development. https://twitter.com/theosanderson/status/885534658091503617">https://twitter.com/theosande...
But what of the mutants that can grow in the blood with a particular gene knocked-out? Do those genes have functions elsewhere in the parasite lifecycle? Could they be required before the parasite even reaches the blood, while it develops hidden inside the liver?
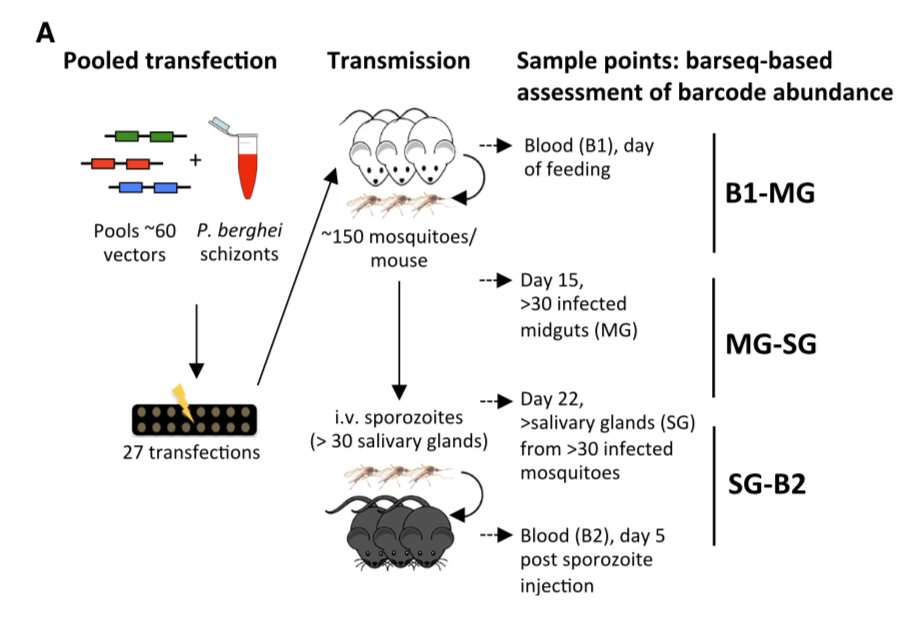
To address this we generated 1,342 mutants in 27 batches at @sangerinstitute. Each pool of parasite was immediately sent to @unibern where it went through an entire parasite lifecycle with samples taken at each stage.
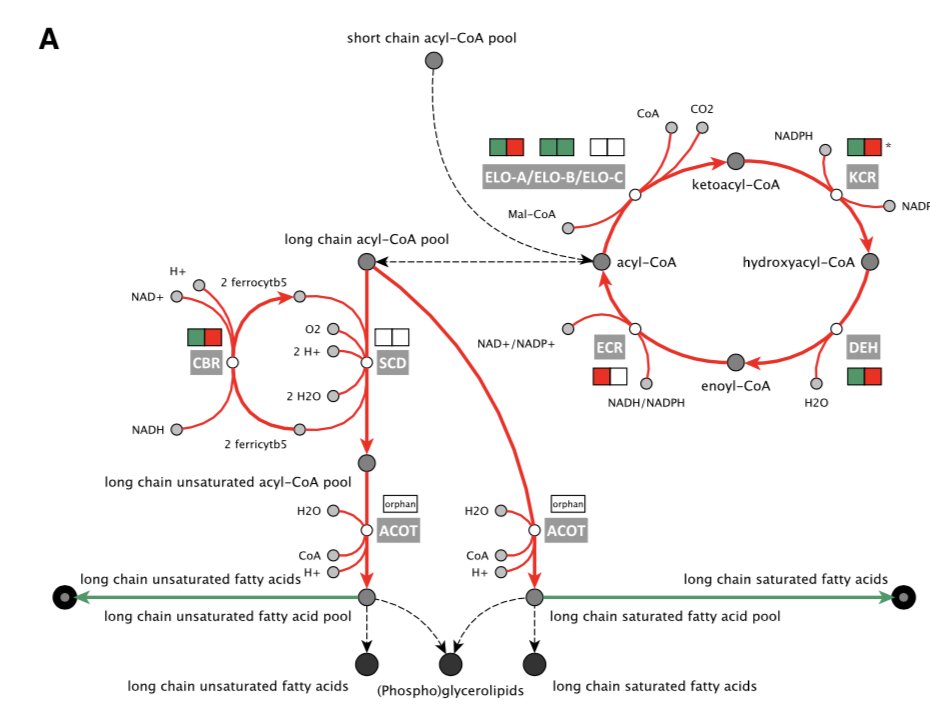
Many genes dispensable in the blood stages didn& #39;t seem to be required in other stages, but a number of pathways stood out for their importance in the liver-stage. For example, in the absence of components of the fatty acid elongation pathway, parasites developed much more slowly:
These results were also captured and predicted by a geome-scale model of P. berghei metabolism in the liver! Lots more gaps to fill with conditional systems, but a substantial step for scaling up our functional-genetic understanding of the harder-to-study stages of malaria.
Huge multi-institutional effort from @unibern @sangerinstitute @realLCSB @LUMCPara @mims_umea. Was great to be part of this!
(led by @rrlimenitakis, @bushell_lab, @AnushChP and Magali Roques - sorry for the fail of only doing Twitter handles)

 Read on Twitter
Read on Twitter