In a paper published this week @NaturePlants, we present the main results of seven years studying #MorningGlory taxonomy and evolution: "A taxonomic monograph of Ipomoea integrated across phylogenetic scales". And there is sweet potato too! https://www.nature.com/articles/s41477-019-0535-4">https://www.nature.com/articles/...  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🍠" title="Roasted sweet potato" aria-label="Emoji: Roasted sweet potato">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🍠" title="Roasted sweet potato" aria-label="Emoji: Roasted sweet potato"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Downwards arrow" aria-label="Emoji: Downwards arrow">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Downwards arrow" aria-label="Emoji: Downwards arrow"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Downwards arrow" aria-label="Emoji: Downwards arrow">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Downwards arrow" aria-label="Emoji: Downwards arrow">
This project, led by Robert Scotland at @OxfordPlants, involved researchers from @OregonState, @Cipotato, @DukeU, @TheBotanics and @KewScience, as well as really important collaborators during field work, herbarium curators, other taxonomists, funders... So much help! https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏" title="Person with folded hands" aria-label="Emoji: Person with folded hands">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏" title="Person with folded hands" aria-label="Emoji: Person with folded hands">
Our aim was to show how powerful evolutionary studies become when they integrate with a good, comprehensive taxonomic study (especially for relatively poorly known groups of tropical plants). This idea was proposed 7 years ago ( https://www.cell.com/trends/ecology-evolution/fulltext/S0169-5347(12)00101-2)">https://www.cell.com/trends/ec... and here we show it works.
We studied specimens of Ipomoea and related genera (tribe Ipomoeeae, family Convolvulaceae) from all around the world. This, of course, would have been just impossible without an extraordinary resource: HERBARIUM COLLECTIONS!
We obtained DNA from specimens collected during the last 250 years and inferred phylogenies to investigate the evolutionary relationships between species. And guess what? Without taxonomy, 40% specimens in our study would be wrongly identified ( https://abs.twimg.com/emoji/v2/... draggable="false" alt="👁️" title="Eye" aria-label="Emoji: Eye">Fig. 3b). https://www.nature.com/articles/s41477-019-0535-4/figures/3">https://www.nature.com/articles/...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👁️" title="Eye" aria-label="Emoji: Eye">Fig. 3b). https://www.nature.com/articles/s41477-019-0535-4/figures/3">https://www.nature.com/articles/...
Also, check Fig. 3a highlighting & #39;Ipomoea& #39; species present in Bolivia. A comprehensive study of the entire genus allowed us to identify their close relatives, even if they do not appear in Bolivia.
https://www.nature.com/articles/s41477-019-0535-4/figures/3">https://www.nature.com/articles/... (Photo: & #39;Ipomoea santacrucensis& #39;, by John Wood)
https://www.nature.com/articles/s41477-019-0535-4/figures/3">https://www.nature.com/articles/... (Photo: & #39;Ipomoea santacrucensis& #39;, by John Wood)
In total, in this project we described 63 new species (10% species of Ipomoea are new!), identified a lot of synonyms (69% rate), lectotypified almost 300 names, published hundreds of descriptions and illustrations and some very useful identification keys. Massive improvement! https://abs.twimg.com/emoji/v2/... draggable="false" alt="😄" title="Smiling face with open mouth and smiling eyes" aria-label="Emoji: Smiling face with open mouth and smiling eyes">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="😄" title="Smiling face with open mouth and smiling eyes" aria-label="Emoji: Smiling face with open mouth and smiling eyes">
Ah, don& #39;t miss the SPECIES NARRATIVES in the Supplementary Information! We provide ten examples describing how the species discovery process works. Really nice piece to get a sense of what it takes to discover new species.  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🏦" title="Bank" aria-label="Emoji: Bank">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🏦" title="Bank" aria-label="Emoji: Bank"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">
In terms of evolution, we identified two very diverse American clades with high diversification rates, comparable to some of the most iconic evolutionary radiations in the #PlantKingdom. In the picture, some members of this group (described @KewBulletin: https://link.springer.com/article/10.1007/s12225-015-9592-7).">https://link.springer.com/article/1...
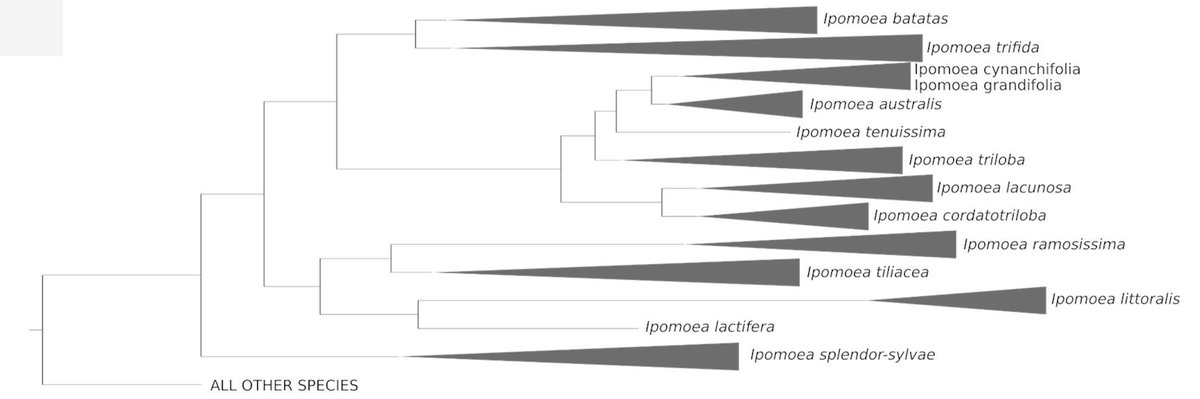
And, of course, there is #SweetPotato! Important results appeared last year in @CurrentBiology ( https://www.cell.com/current-biology/supplemental/S0960-9822(18)30321-X),">https://www.cell.com/current-b... e.g. we published a complete phylogeny of all sweet potato close relatives and identified & #39;Ipomoea trifida& #39; as its closest relative.
Now, we show that:
1) Both species diverged over 1 MYA.
2) Part of the diversity existing within the crop largely predates agriculture.
3) At least 60 other Ipomoea species evolved storage roots over 1 MYA.
4) Many of them are as big as sweet potatoes & edible ( https://abs.twimg.com/emoji/v2/... draggable="false" alt="📷" title="Camera" aria-label="Emoji: Camera">I. lilloana)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="📷" title="Camera" aria-label="Emoji: Camera">I. lilloana)
SO:
1) Both species diverged over 1 MYA.
2) Part of the diversity existing within the crop largely predates agriculture.
3) At least 60 other Ipomoea species evolved storage roots over 1 MYA.
4) Many of them are as big as sweet potatoes & edible (
SO:
The storage root in cultivated #sweetpotato is NOT a product of human domestication but rather an existing trait that predisposed the plant for cultivation.
What changes did human domestication produce in the crop then? We will try to answer this question in forthcoming studies.
What changes did human domestication produce in the crop then? We will try to answer this question in forthcoming studies.
In conclusion: we can make extraordinary progress in our understanding of world& #39;s #biodiversity by integrating specimen-based taxonomy with genomics and phylogenetics. The resources (Natural History collections!) and scientific expertise are already available. Let& #39;s do it!  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌎" title="Earth globe americas" aria-label="Emoji: Earth globe americas">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌎" title="Earth globe americas" aria-label="Emoji: Earth globe americas">
PS: for those wondering where the ACTUAL taxonomic monograph is, stay tuned. It has been already accepted for publication and will be out soon, full of descriptions, images, identification keys... https://abs.twimg.com/emoji/v2/... draggable="false" alt="😄" title="Smiling face with open mouth and smiling eyes" aria-label="Emoji: Smiling face with open mouth and smiling eyes">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="😄" title="Smiling face with open mouth and smiling eyes" aria-label="Emoji: Smiling face with open mouth and smiling eyes">

 Read on Twitter
Read on Twitter " title="This project, led by Robert Scotland at @OxfordPlants, involved researchers from @OregonState, @Cipotato, @DukeU, @TheBotanics and @KewScience, as well as really important collaborators during field work, herbarium curators, other taxonomists, funders... So much help!https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏" title="Person with folded hands" aria-label="Emoji: Person with folded hands">" class="img-responsive" style="max-width:100%;"/>
" title="This project, led by Robert Scotland at @OxfordPlants, involved researchers from @OregonState, @Cipotato, @DukeU, @TheBotanics and @KewScience, as well as really important collaborators during field work, herbarium curators, other taxonomists, funders... So much help!https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏" title="Person with folded hands" aria-label="Emoji: Person with folded hands">" class="img-responsive" style="max-width:100%;"/>


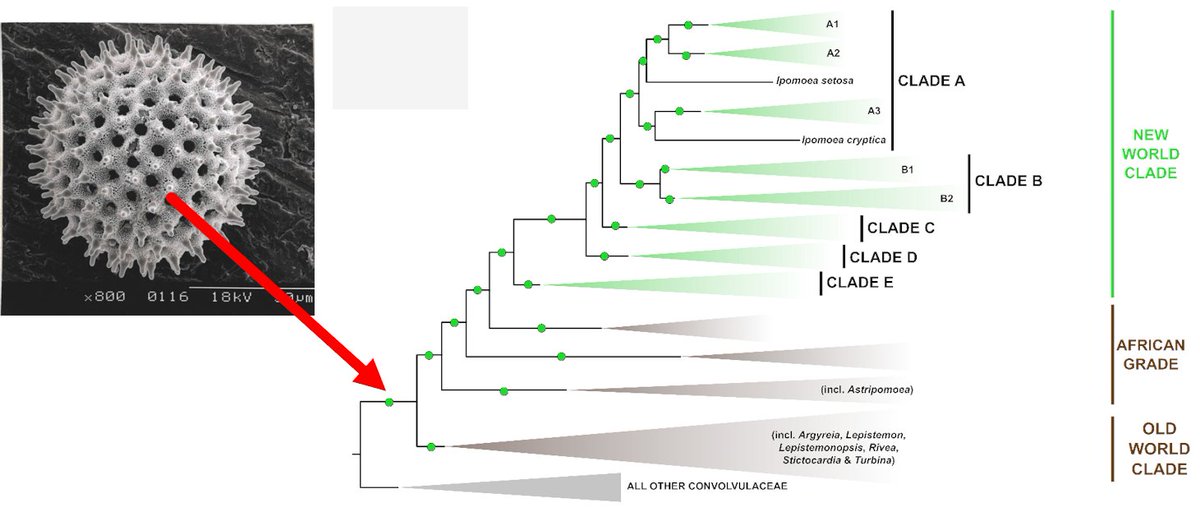 IMPORTANT TAXONOMIC RESULT: we expand Ipomoea to include all members of the tribe Ipomoeeae (Convolvulaceae with spiny pollen). Prev. studies suggested non-monophyly of other previously recognised genera & we confirmed it. All nomenclatural changes here: https://static-content.springer.com/esm/art%3..." title="https://abs.twimg.com/emoji/v2/... draggable="false" alt="🚩" title="Triangular flag on post" aria-label="Emoji: Triangular flag on post">IMPORTANT TAXONOMIC RESULT: we expand Ipomoea to include all members of the tribe Ipomoeeae (Convolvulaceae with spiny pollen). Prev. studies suggested non-monophyly of other previously recognised genera & we confirmed it. All nomenclatural changes here: https://static-content.springer.com/esm/art%3..." class="img-responsive" style="max-width:100%;"/>
IMPORTANT TAXONOMIC RESULT: we expand Ipomoea to include all members of the tribe Ipomoeeae (Convolvulaceae with spiny pollen). Prev. studies suggested non-monophyly of other previously recognised genera & we confirmed it. All nomenclatural changes here: https://static-content.springer.com/esm/art%3..." title="https://abs.twimg.com/emoji/v2/... draggable="false" alt="🚩" title="Triangular flag on post" aria-label="Emoji: Triangular flag on post">IMPORTANT TAXONOMIC RESULT: we expand Ipomoea to include all members of the tribe Ipomoeeae (Convolvulaceae with spiny pollen). Prev. studies suggested non-monophyly of other previously recognised genera & we confirmed it. All nomenclatural changes here: https://static-content.springer.com/esm/art%3..." class="img-responsive" style="max-width:100%;"/>
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">" title="Ah, don& #39;t miss the SPECIES NARRATIVES in the Supplementary Information! We provide ten examples describing how the species discovery process works. Really nice piece to get a sense of what it takes to discover new species. https://abs.twimg.com/emoji/v2/... draggable="false" alt="🏦" title="Bank" aria-label="Emoji: Bank">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">" title="Ah, don& #39;t miss the SPECIES NARRATIVES in the Supplementary Information! We provide ten examples describing how the species discovery process works. Really nice piece to get a sense of what it takes to discover new species. https://abs.twimg.com/emoji/v2/... draggable="false" alt="🏦" title="Bank" aria-label="Emoji: Bank">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">">
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">" title="Ah, don& #39;t miss the SPECIES NARRATIVES in the Supplementary Information! We provide ten examples describing how the species discovery process works. Really nice piece to get a sense of what it takes to discover new species. https://abs.twimg.com/emoji/v2/... draggable="false" alt="🏦" title="Bank" aria-label="Emoji: Bank">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">" title="Ah, don& #39;t miss the SPECIES NARRATIVES in the Supplementary Information! We provide ten examples describing how the species discovery process works. Really nice piece to get a sense of what it takes to discover new species. https://abs.twimg.com/emoji/v2/... draggable="false" alt="🏦" title="Bank" aria-label="Emoji: Bank">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔬" title="Microscope" aria-label="Emoji: Microscope">https://abs.twimg.com/emoji/v2/... draggable="false" alt="📚" title="Books" aria-label="Emoji: Books">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌿" title="Herb" aria-label="Emoji: Herb">">


 I. lilloana)SO:" title="Now, we show that:1) Both species diverged over 1 MYA.2) Part of the diversity existing within the crop largely predates agriculture.3) At least 60 other Ipomoea species evolved storage roots over 1 MYA.4) Many of them are as big as sweet potatoes & edible (https://abs.twimg.com/emoji/v2/... draggable="false" alt="📷" title="Camera" aria-label="Emoji: Camera">I. lilloana)SO:" class="img-responsive" style="max-width:100%;"/>
I. lilloana)SO:" title="Now, we show that:1) Both species diverged over 1 MYA.2) Part of the diversity existing within the crop largely predates agriculture.3) At least 60 other Ipomoea species evolved storage roots over 1 MYA.4) Many of them are as big as sweet potatoes & edible (https://abs.twimg.com/emoji/v2/... draggable="false" alt="📷" title="Camera" aria-label="Emoji: Camera">I. lilloana)SO:" class="img-responsive" style="max-width:100%;"/>


