I& #39;m going to try something new - here& #39;s a thread of the talk I gave at #WSN2019 with @Matt_Leray. The talk was about the diet of the giant plumose anemone ud835ude14ud835ude26ud835ude35ud835ude33ud835ude2aud835ude25ud835ude2aud835ude36ud835ude2e ud835ude27ud835ude22ud835ude33ud835ude24ud835ude2aud835ude2eud835ude26ud835ude2f and how it compared to the plankton community. Please ask questions!
PC: T. Dwyer.
1/n
PC: T. Dwyer.
1/n
You may be asking: "Why should I care about the diet of anemones?" Well, for one, it& #39;s eating some of the things you may like to eat: fishes, clams, and crabs to name a few. They can dominate a system and have been shown to beat out most other things attached to the bottom.
2/n
2/n
The bottom of the ocean, especially in the shallows, is dominated by predators consuming the prey floating by (plankton) and can have major impacts on the plankton. Several studies found that this community can completely deplete the nearby floating microalgae.
PC: T. Dwyer
3/n
PC: T. Dwyer
3/n
That& #39;s not to say that plankton can& #39;t defend themselves. They can be toxic, have standing defenses, or avoid predators. This thread is all about prey selectivity: the consumption of prey in a ratio different from available prey.
PC: P. Bryant - http://nathistoc.bio.uci.edu/crustacea/Larvae/Larva1.htm
4/n">https://nathistoc.bio.uci.edu/crustacea...
PC: P. Bryant - http://nathistoc.bio.uci.edu/crustacea/Larvae/Larva1.htm
4/n">https://nathistoc.bio.uci.edu/crustacea...
Why do these types of study matter? They help us understand the effects of predators on their ecosystem, how prey are captured, and how predators partition the available prey. Unfortunately, quantifying the diet of animals that eat plankton is difficult.
5/n
5/n
Plankton are frequently microscopic and soft-bodied plankton are digested quicker than hard-bodied ones (think  https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc1b" title="Bug" aria-label="Emoji: Bug">v
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc1b" title="Bug" aria-label="Emoji: Bug">v https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc0c" title="Snail" aria-label="Emoji: Snail">). This makes it difficult to identify plankton beyond the most broad of groups using traditional diet analyses.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc0c" title="Snail" aria-label="Emoji: Snail">). This makes it difficult to identify plankton beyond the most broad of groups using traditional diet analyses.
PC: B. Vellutini
6/n
PC: B. Vellutini
6/n
For those that aren& #39;t aware of how one would traditionally examine gut contents, here& #39;s the recipe for working with anemones: Collect an anemone, ice it, cut it in half  https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dude2d" title="Loudly crying face" aria-label="Emoji: Loudly crying face">, scrape out the contents, rinse with LOTS of alcohol (for the mucus and preservation), ID everything.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dude2d" title="Loudly crying face" aria-label="Emoji: Loudly crying face">, scrape out the contents, rinse with LOTS of alcohol (for the mucus and preservation), ID everything.
7/n
7/n
In this dish of gut contents, you can see an amphipod (relative of  https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd9e" title="Lobster" aria-label="Emoji: Lobster">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd9e" title="Lobster" aria-label="Emoji: Lobster"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd90" title="Shrimp" aria-label="Emoji: Shrimp">+
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd90" title="Shrimp" aria-label="Emoji: Shrimp">+ https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd80" title="Crab" aria-label="Emoji: Crab">) circled. There& #39;s a whole lot of anemone tissue also. With new molecular techniques like DNA metabarcoding, we can analyze diets a lot faster, more cheaply, and with less need for taxonomic expertise.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd80" title="Crab" aria-label="Emoji: Crab">) circled. There& #39;s a whole lot of anemone tissue also. With new molecular techniques like DNA metabarcoding, we can analyze diets a lot faster, more cheaply, and with less need for taxonomic expertise.
8/n
8/n
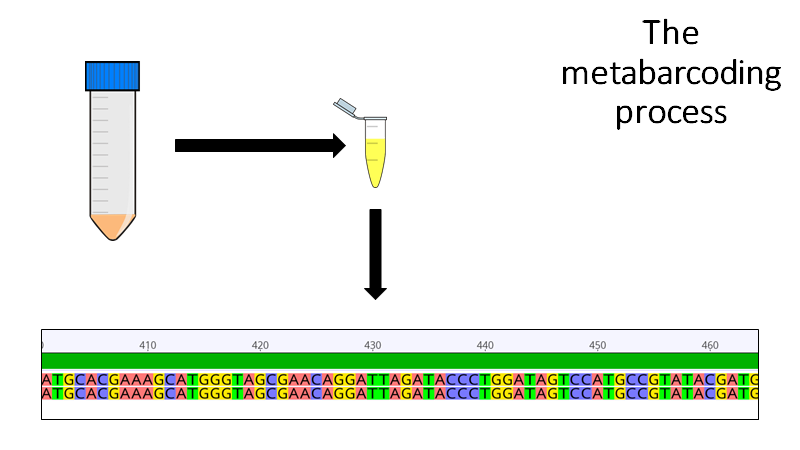
To metabarcode a sample, all you need to do is collect a community sample, grind it all up, extract the DNA, send that DNA off to a sequencing center, and then match those sequences to previously collected sequences to identify what& #39;s in the gut contents.
9/n
9/n
We used metabarcoding to compare anemone diets to the available plankton. Plankton was collected with two mesh sizes: 80 um (nearly everything) and 330 um (big things). We wanted to know, ud835ude04ud835uddf5ud835uddf6ud835uddf0ud835uddf5 ud835uddfcud835uddf3 ud835ude01ud835uddf5ud835uddf2ud835ude00ud835uddf2 ud835ude04ud835uddeeud835ude00 ud835uddfaud835uddfcud835uddffud835uddf2 ud835uddf9ud835uddf6ud835uddf8ud835uddf2 ud835ude01ud835uddf5ud835uddf2 ud835uddeeud835uddfbud835uddf2ud835uddfaud835uddfcud835uddfbud835uddf2& #39;ud835ude00 ud835uddf1ud835uddf6ud835uddf2ud835ude01.
10/n
10/n
This work was all done at Friday Harbor Labs ( @MarineBiol_FHL ) as part of my PhD work. FHL is located on the San Juan Islands in the Salish Sea in Washington and is a wonderful place to learn about the field of invertebrate zoology, amongst other fields.
PC: J. Mabel
11/n
PC: J. Mabel
11/n
We had two questions for this project: ud835uddd7ud835uddfcud835uddf2ud835ude00 ud835ude01ud835uddf5ud835uddf2 ud835uddf4ud835uddf6ud835uddeeud835uddfbud835ude01 ud835uddfdud835uddf9ud835ude02ud835uddfaud835uddfcud835ude00ud835uddf2 ud835uddeeud835uddfbud835uddf2ud835uddfaud835uddfcud835uddfbud835uddf2 ud835uddf5ud835uddeeud835ude03ud835uddf2 ud835uddee ud835ude00ud835uddf2ud835uddf9ud835uddf2ud835uddf0ud835ude01ud835uddf6ud835ude03ud835uddf2 ud835uddf1ud835uddf6ud835uddf2ud835ude01 ud835uddeeud835uddfbud835uddf1 ud835uddf5ud835uddfcud835ude04 ud835uddf1ud835uddfcud835uddf2ud835ude00 ud835uddfaud835uddf2ud835ude01ud835uddeeud835uddefud835uddeeud835uddffud835uddf0ud835uddfcud835uddf1ud835uddf6ud835uddfbud835uddf4 ud835uddf0ud835uddfcud835uddfaud835uddfdud835uddeeud835uddffud835uddf2 ud835ude01ud835uddfc ud835ude01ud835uddffud835uddeeud835uddf1ud835uddf6ud835ude01ud835uddf6ud835uddfcud835uddfbud835uddeeud835uddf9 ud835uddf4ud835ude02ud835ude01 ud835uddf0ud835uddfcud835uddfbud835ude01ud835uddf2ud835uddfbud835ude01 ud835uddeeud835uddfbud835uddeeud835uddf9ud835ude06ud835ude00ud835uddf6ud835ude00?
12/n
12/n
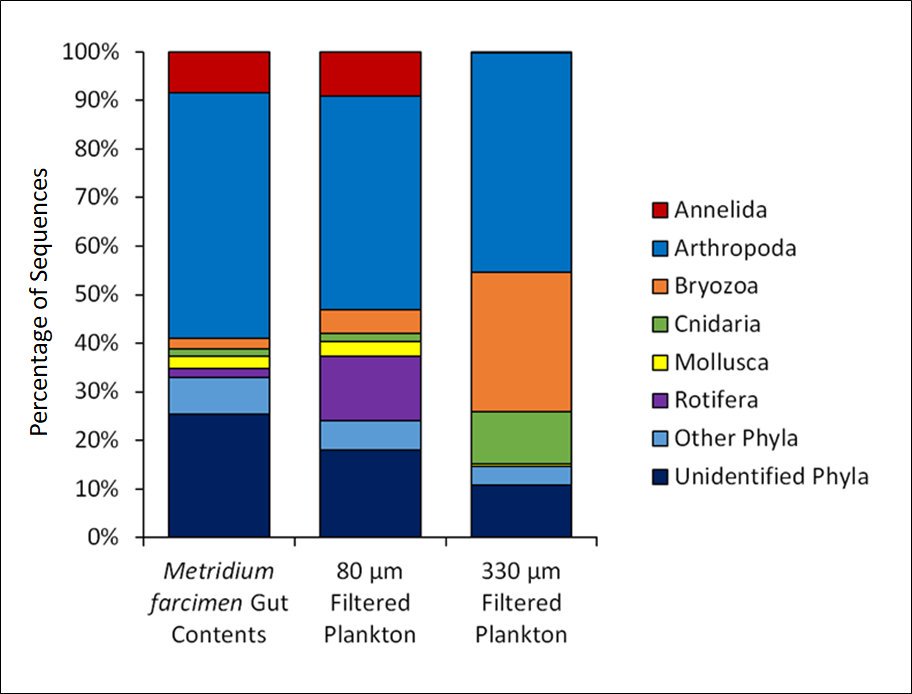
Now for some results. You can see that there is ud835uddee ud835ude00ud835ude01ud835uddffud835uddfcud835uddfbud835uddf4 ud835ude00ud835uddf6ud835uddfaud835uddf6ud835uddf9ud835uddeeud835uddffud835uddf6ud835ude01ud835ude06 ud835uddf6ud835uddfb ud835ude01ud835uddf5ud835uddf2 ud835uddf0ud835uddfcud835uddfaud835uddfdud835uddfcud835ude00ud835uddf6ud835ude01ud835uddf6ud835uddfcud835uddfb ud835uddefud835uddf2ud835ude01ud835ude04ud835uddf2ud835uddf2ud835uddfb ud835ude01ud835uddf5ud835uddf2 ud835uddeeud835uddfbud835uddf2ud835uddfaud835uddfcud835uddfbud835uddf2 ud835uddf4ud835ude02ud835ude01ud835ude00 ud835uddeeud835uddfbud835uddf1 ud835ude01ud835uddf5ud835uddf2 ud835udff4ud835udfec ud835ude02ud835uddfa ud835uddf3ud835uddf6ud835uddf9ud835ude01ud835uddf2ud835uddffud835uddf2ud835uddf1 ud835uddfdud835uddf9ud835uddeeud835uddfbud835uddf8ud835ude01ud835uddfcud835uddfb and less between the 330 and gut contents.
13/n
13/n
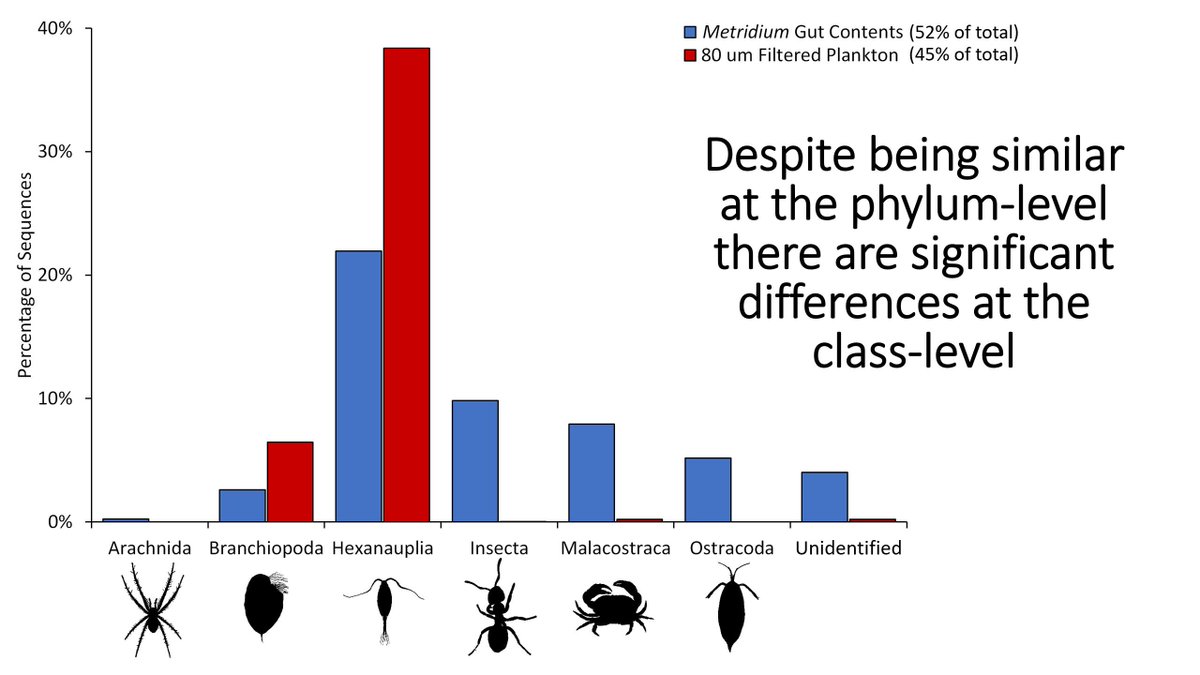
If we break up the Arthropoda group and look at subgroups, the differences between the plankton and the anemones becomes more apparent. Blue bars: anemone diets; red bars: plankton. There are some large differences you can see here (including 9% of the diet being insects)!
14/n
14/n
The DNA for these insects matches the yellow meadow ant which, at the time of this work, was releasing winged queens and drones, some of which likely ended up in the water. This is an interesting link between the terrestrial and marine environment!
PC: Getty
15/n
PC: Getty
15/n
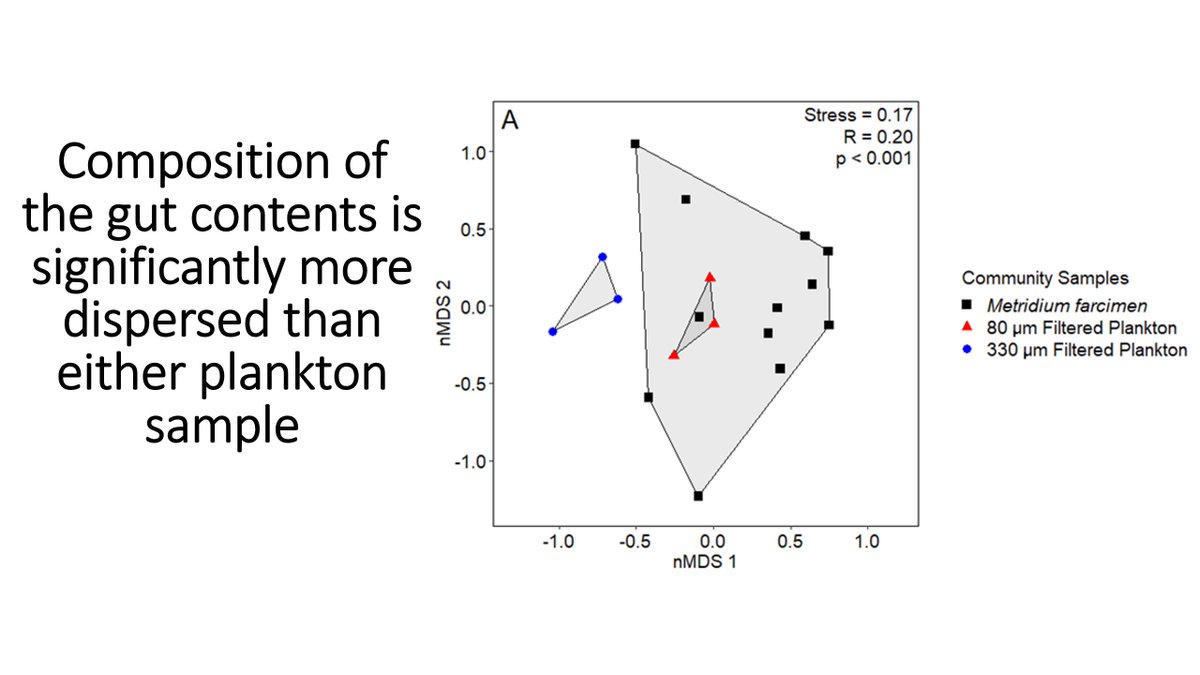
Here& #39;s an nMDS plot. While the nem gut contents (GC) overlap the 80 um plankton samples (indicates similarity), the GC are more dispersed - ud835uddfbud835uddf2ud835uddfaud835ude00 ud835uddf0ud835uddeeud835ude01ud835uddf0ud835uddf5 ud835uddfdud835uddffud835uddf2ud835ude06 ud835uddfcud835ude03ud835uddf2ud835uddff ud835uddf5ud835uddffud835ude00 ud835ude04ud835uddf5ud835uddf6ud835uddf9ud835uddf2 ud835uddfdud835uddf9ud835uddeeud835uddfbud835uddf8ud835ude01ud835uddfcud835uddfb ud835ude00ud835uddeeud835uddfaud835uddfdud835uddf9ud835uddf2ud835ude00 ud835uddeeud835uddffud835uddf2 ud835uddee ud835ude00ud835uddfbud835uddeeud835uddfdud835ude00ud835uddf5ud835uddfcud835ude01 ud835uddf6ud835uddfb ud835ude01ud835uddf6ud835uddfaud835uddf2.
16/n
16/n
When compared to either plankton sampling, there were many species that were significantly less abundant in the gut contents (pref. avoidance or can& #39;t be captured), but no significantly more abundant (no. pref. consumption).
17/n
17/n
What this means is ud835ude01ud835uddf5ud835uddf2ud835uddffud835uddf2 ud835uddf6ud835ude00 ud835uddfdud835uddffud835uddf2ud835ude06 ud835ude00ud835uddf2ud835uddf9ud835uddf2ud835uddf0ud835ude01ud835uddf6ud835ude03ud835uddf6ud835ude01ud835ude06 ud835uddf6ud835uddfb ud835ude01ud835uddf5ud835uddf2 ud835uddf4ud835uddf6ud835uddeeud835uddfbud835ude01 ud835uddfdud835uddf9ud835ude02ud835uddfaud835uddfcud835ude00ud835uddf2 ud835uddeeud835uddfbud835uddf2ud835uddfaud835uddfcud835uddfbud835uddf2; they seem to consume all prey that they can capture, missing species that are too small (like rotifers) and good swimmers (like copepods).
PC: Kils
18/n
PC: Kils
18/n
Now let& #39;s compare traditional techniques to metabarcoding. Columns are different studies and shaded boxes indicate that that group was present. There& #39;s a much higher diversity of groups found using the metabarcoding approach (26 v 7 groups).
19/n
19/n
TL;DR:
1. There was prey selectivity
2. Diet was similar to the 80 um plankton samples
3. Surprising amount of terrestrial input ( https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc1c" title="Ant" aria-label="Emoji: Ant">) in the diet
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc1c" title="Ant" aria-label="Emoji: Ant">) in the diet
4. Metabarcoding was an excellent approach for examining the diversity of prey in a plankton predator
20/n
1. There was prey selectivity
2. Diet was similar to the 80 um plankton samples
3. Surprising amount of terrestrial input (
4. Metabarcoding was an excellent approach for examining the diversity of prey in a plankton predator
20/n
This work was done with funding from @AMNH, @UWBiology, and @MarineBiol_FHL. We had lots of lab and field help from @chiton_chitoff, @thefishbotherer, and C. Alexandra.
Thanks to @WSN_Secretariat for the conference and thank you for taking a read!
Thanks to @WSN_Secretariat for the conference and thank you for taking a read!

 Read on Twitter
Read on Twitter



 vhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc0c" title="Snail" aria-label="Emoji: Snail">). This makes it difficult to identify plankton beyond the most broad of groups using traditional diet analyses.PC: B. Vellutini6/n" title="Plankton are frequently microscopic and soft-bodied plankton are digested quicker than hard-bodied ones (think https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc1b" title="Bug" aria-label="Emoji: Bug">vhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc0c" title="Snail" aria-label="Emoji: Snail">). This makes it difficult to identify plankton beyond the most broad of groups using traditional diet analyses.PC: B. Vellutini6/n" class="img-responsive" style="max-width:100%;"/>
vhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc0c" title="Snail" aria-label="Emoji: Snail">). This makes it difficult to identify plankton beyond the most broad of groups using traditional diet analyses.PC: B. Vellutini6/n" title="Plankton are frequently microscopic and soft-bodied plankton are digested quicker than hard-bodied ones (think https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc1b" title="Bug" aria-label="Emoji: Bug">vhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83dudc0c" title="Snail" aria-label="Emoji: Snail">). This makes it difficult to identify plankton beyond the most broad of groups using traditional diet analyses.PC: B. Vellutini6/n" class="img-responsive" style="max-width:100%;"/>
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd90" title="Shrimp" aria-label="Emoji: Shrimp">+https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd80" title="Crab" aria-label="Emoji: Crab">) circled. There& #39;s a whole lot of anemone tissue also. With new molecular techniques like DNA metabarcoding, we can analyze diets a lot faster, more cheaply, and with less need for taxonomic expertise.8/n" title="In this dish of gut contents, you can see an amphipod (relative of https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd9e" title="Lobster" aria-label="Emoji: Lobster">https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd90" title="Shrimp" aria-label="Emoji: Shrimp">+https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd80" title="Crab" aria-label="Emoji: Crab">) circled. There& #39;s a whole lot of anemone tissue also. With new molecular techniques like DNA metabarcoding, we can analyze diets a lot faster, more cheaply, and with less need for taxonomic expertise.8/n" class="img-responsive" style="max-width:100%;"/>
https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd90" title="Shrimp" aria-label="Emoji: Shrimp">+https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd80" title="Crab" aria-label="Emoji: Crab">) circled. There& #39;s a whole lot of anemone tissue also. With new molecular techniques like DNA metabarcoding, we can analyze diets a lot faster, more cheaply, and with less need for taxonomic expertise.8/n" title="In this dish of gut contents, you can see an amphipod (relative of https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd9e" title="Lobster" aria-label="Emoji: Lobster">https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd90" title="Shrimp" aria-label="Emoji: Shrimp">+https://abs.twimg.com/emoji/v2/... draggable="false" alt="ud83eudd80" title="Crab" aria-label="Emoji: Crab">) circled. There& #39;s a whole lot of anemone tissue also. With new molecular techniques like DNA metabarcoding, we can analyze diets a lot faster, more cheaply, and with less need for taxonomic expertise.8/n" class="img-responsive" style="max-width:100%;"/>